MicroRNA: A Major Key in Pain Neurobiology- Juniper Publishers
Juniper Publisher- Journal of Cell science
Abstract
MiRNAs are single-stranded small noncoding RNAs that
consist of approximately 22 nucleotides, that are involve in a wide
range of biological processes including pain physiopathology. Because of
their role as master switches in regulation and signaling pathways
through modifications in nociceptive receptors, ion channels,
pro-inflammatory molecules, emotional and cognitional behaviors
associated with pain, the triggered enthusiasm for miRNAs as promising
therapeutic targets is still active. Furthermore, the expression of
specific miRNA can be helpful to predict treatment response in patients
which suffer pain conditions that are poorly controlled by the currently
available analgesics. This evidence is supported in several researches
with animal pain models that we briefly review in this article to
approximate in the understand of the role and neurobiology process
through miRNA represents a major key for future therapeutics in pain,
emphasizing in the neuropathic pain condition.
Keywords: MicroRNA; Neurobiology; Neuropathic painIntroduction
Neuropathic pain (NP) represents one of main causes
of chronic pain, perhaps trailing only osteoarthritis as a cause [1].
One of the keys to understand the biology of neuropathic pain is to know
that´s is caused by an injury in a nervous tissue and that nociceptive
pathways are involve in the lesion [2]. In the clinical practice, the
most frequent NP origins are the neurons of dorsal root ganglia (DRG)
and trigeminal ganglia (TG) in mechanical, metabolic and toxic lesions
as traumatic injury, herpes zoster, diabetes, or cancer chemotherapy,
all this kind of pathologies promotes functional changes in the
initiation and maintenance of NP [3,4].
The several changes observed in neuropathic pain
condition are well represented in two major symptoms, allodynia and
hyperalgesia. Both symptoms are observed in patients and as signs in
animal models of chronic pain such as the spinal nerve ligation (SNL),
consisting in a tight ligation of L5 and L6 spinal nerves were Fukuoka
et al. [5] described a down-regulation of the inhibitory γ-amino butyric
acid receptor A (GABAA) in the dorsal root ganglia (DRG). In spared
nerve injury (SNI) were shown an up-regulation of interleukin-1β (IL-1β)
in the prefrontal cortex of rats [6]. And many other changes can be
observe in every pre-clinical model of pain which includes up-regulation
of interleukin-6 (IL-6) [7], neurokinin-1 receptor in the dorsal horn
[8], down regulation of dopaminergic D1 and D2 receptors in the
anterior cingulate cortex in a rat model [9] just to mention a few.
Clearly, this changes in the substances and receptors regulation
are product of an altered gene expression in the nociceptive
pathways. One of the most recent studied mechanisms that explains the
pathogenesis and play a crucial role in fine-tuning gene expression [10]
in the chronic pain is MicroRNAs (miRNAs) regulation, that are involve
in a wide range of biological processes [11]. In this review we will
focus in the role and neurobiology process through miRNA represents a
major key for future therapeutics in pain, emphasizing in the NP
condition.
Biology and mechanisms of miRNA
MiRNAs are single-stranded small noncoding RNAs that
consist of approximately 22 nucleotides. The genomic location of miRNAs
can be broadly divided into intergenic (between genes) or intronic
(embedded into a gene) [11]. After the transcription of a coding DNA
protein is expressed the precursor messenger RNA (pre-mRNA) which
conformation includes 4 regions, 5′-untranslated region (UTR), the
protein-coding exon, the noncoding intron, and the 3′-UTR, that
determines the main targets of miRNA [12]. The intronic or
intron-derived microRNA (Id-miRNA) is formed in the in-frame introns and
the intergenic miRNAs are set between independent transcription units
[13], both has the capability of degrading messenger-RNA (mRNA) and
inhibit protein translation so they share not only functional but also
structural properties. With the only difference that intronic miRNA are
typically transcribed from the same promoter as their host genes (Pol
II) and require RNA splicing machinery [14-16] while intergenic RNAs
genes have their own transcription regulatory elements [13].
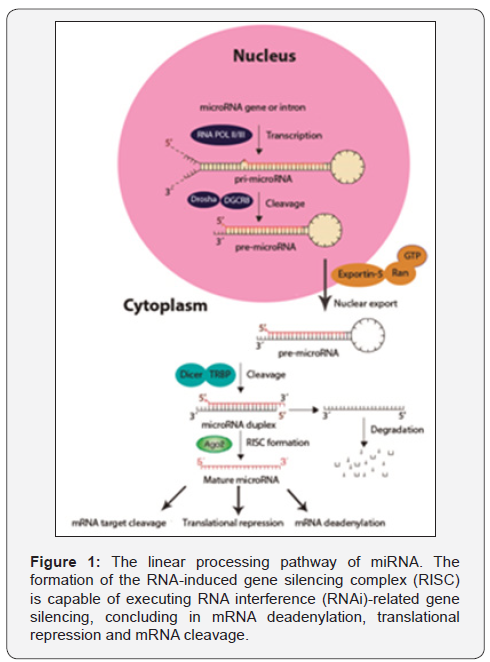
In Figure 1 is represented the genesis and mechanism by
which the interaction between miRNA, the target mRNA and the
RNA-induced gene silencing complex (RISC) suppress the gene
expression. This process begin with the excision of the primary
precursor microRNA (pri-miRNA) by the RNA polymerase type-
II (Pol-II) [17], this pri-miRNA at certain concentration can make
a negative feedback to Pol-II. Then if the pri-miRNA is origin in
an exon, it will be cropped into the hairpin-shaped pre-miRNAs
by nuclear RNase III Drosha [18] or by spliceosomal components
if comes from introns to form a mature precursor miRNA (premiRNA).
This pre-miRNA is exported out of the nucleus to the
cytoplasm by a member of a Ran-dependent nuclear transport
receptor family, the exportin-5 (Exp5) [19] where is cleaved to
the Dicer-like nucleases to form mature miRNA [20]. Finally the
miRNA is coupled to a ribo nuclear particle (RNP) to get the
RISC which is capable of executing RNA interference (RNAi)-
related gene silencing, concluding in the inhibition of the protein
translation [21].
MiRNA and Pain
The comprehension of the extensive pathways involved in
the genesis of pain put in evidence that the genetic basis play a
major role in pain biology [22]. In the very last years the focus
of researches have been in looking not in an specific target or
individual receptor but instead in a “major switch” that would
regulate multiple gene products and orchestrate multiple
pathways [23] and the recent evidence propose miRNA to be
that switch. The miRNAs have been implied in inflammation [24]
process and other pain conditions such as neuropathic pain [25]
and fibromyalgia [26]. This both common clinical problems are
usually poorly controlled by the currently available analgesics
[27], the reason might be the complex and multiple processing
of nociceptive information in pathological conditions [28].
The changes in this processing are the cause of phenomes like
hyperexcitability that can be induced by a posttranslational
modulation of ion channels, such as voltage-gated sodium
channels [29] or long term potentiation (LTP) and disinhibition
that are product of synaptic modifications [30]. So, this phenomes
initiated by altered processing in nociceptive pathways respond
to certain structures like spinal glial cells, especially microglia
and astrocytes that also plays a major role in pain modulation
[31] and can be govern by epigenetic mechanisms such as DNA
methylation, histone modification, and miRNA expressions [32].
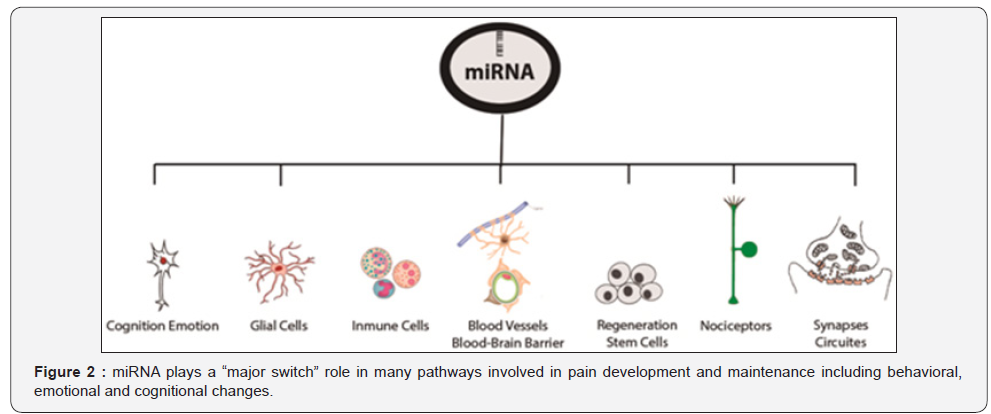
This supports the evidence of the critical role of miRNAs
in pain biology, but not only at molecular, network or synaptic
level, the miRNAs are implied in behavioral, emotional and
cognitional changes [33] that affects pain perception [34]
(Figure 2). However, the expression of miRNAs in DRG, spinal
cord, and brain regions such as the limbic system and prefrontal
cortex can vary from the different causes of pain [4]. The Table
1, resumes some of the most representative miRNAs expressed
in certain pathologies and animal model of acute and chronic
pain, excluding neuropathic pain that will be considered in the
next section. Figure 2. miRNA plays a “major switch” role in
many pathways involved in pain development and maintenance
including behavioral, emotional and cognitional changes.
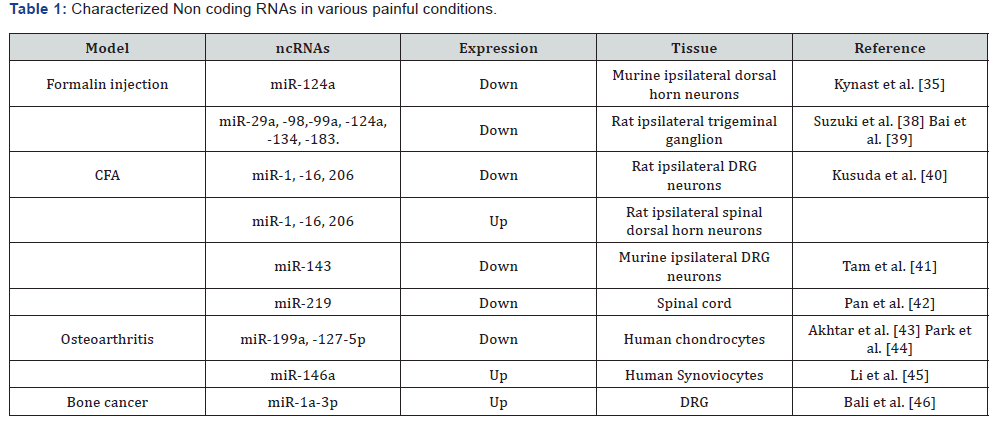
In the case of acute pain, the intra plantar formalin injection,
was shown to decrease miR-124a expression in murine
nociceptive spinal neurons in the ipsilateral horn [35], which
importance seems to be related to the Methyl CpG binding
protein 2 (MeCP2) a multifunctional epigenetic regulator that
is best known for its role in the neurological disorders [36]
and inflammatory pain [37]. Also, the tongue heat hyperalgesia
following complete Freund’s adjuvant (CFA) injection shown
that MeCP2 is involved in regulation of the transient receptor
potential vanilloid 1 (TRPV1) expression in TG neurons [38],
supporting the evidence of the down regulation of miRNA-
124a for the expression of MeCP2. Other works revealed by a
real-time reverse-transcription polymerase chain reaction (RTPCR)
a significant, but differential, downregulation of mature
miR-10a, -29a, -98, -99a, -124a, -134, and -183 in the ipsilateral
mandibular division (V3) of the TG within 4hr after CFA [39], this
down regulation of miRNA releases the translation inhibition of
target mRNAs, thus yielding more proteins that may be relevant
to the development and/or maintenance of inflammatory pain
as Bai et al. [25] conclude. In 2011 Kusuda et al. [40] found that
CFA-induced inflammation significantly reduced miRs-1-16 and
-206 expression in DRG. Conversely, in the spinal dorsal horn all
three miRNAs monitored were up regulated [40]. Tam et al. [41]
demonstrate for the first time that miR-143 expression in DRG
nociceptive neurons is declined in response to inflammation
[41]. More recently, Pan et al. [42] using a CFA model concluded
that methylation-mediated epigenetic modification of spinal
miR-219 expression regulates chronic inflammatory pain by
targeting calcium/calmodulin-dependent protein kinase II
γ (CaMKIIγ) which regulates NMDAR signaling and central
sensitization [42].
In human chondrocytes with IL-1β in vitro stimulation,
revealed that the treatment with p38- mitogen-activated protein
kinase (MAPK) inhibitor (SB202190), enhanced the expression
of miR-199a* which can directly target COX-2 mRNA and reduce
protein expression levels [43]. Considering the IL-1β as a major
mediator in chronic pain, described that miR-127-5p regulates
MMP-13 expression and IL-1β–induced catabolic responses
in human chondrocytes too [44]. Finally, another miRNA, the
miR-146a expressed at reduced levels in DRGs and dorsal
horn of the spinal cords from rats with Osteoarthritis (OA)-
induced pain significantly modulates inflammatory cytokines
and pain-related molecules (e.g. TNFα, COX-2, iNOS, IL-6, IL8,
RANTS and ion channel, TRPV1) [45]. In cancer-associated pain,
another form of chronic pain, miR-1a-3p plays an important role
attenuating the mechanical hypersensitivity [46], however in
this pain condition, it might been implied a large list of miRs.
Role and Expression of miRNAs in Neuropathic pain
The role of miRNA in the regulation of nociception,
endogenous analgesia and in the circuitries and cognitive,
emotional and behavioral components involved in pain is
expected to shed new light on the enigmatic pathophysiology
of neuropathic pain [24]. Therefore disruption of miRNA
processing in primary afferent pathways is sufficient to inhibit
injury-induced long-term development of chronic pain-related
behaviors, this affirmation is supported in a large evidence of
investigations we resumed in Table 2.
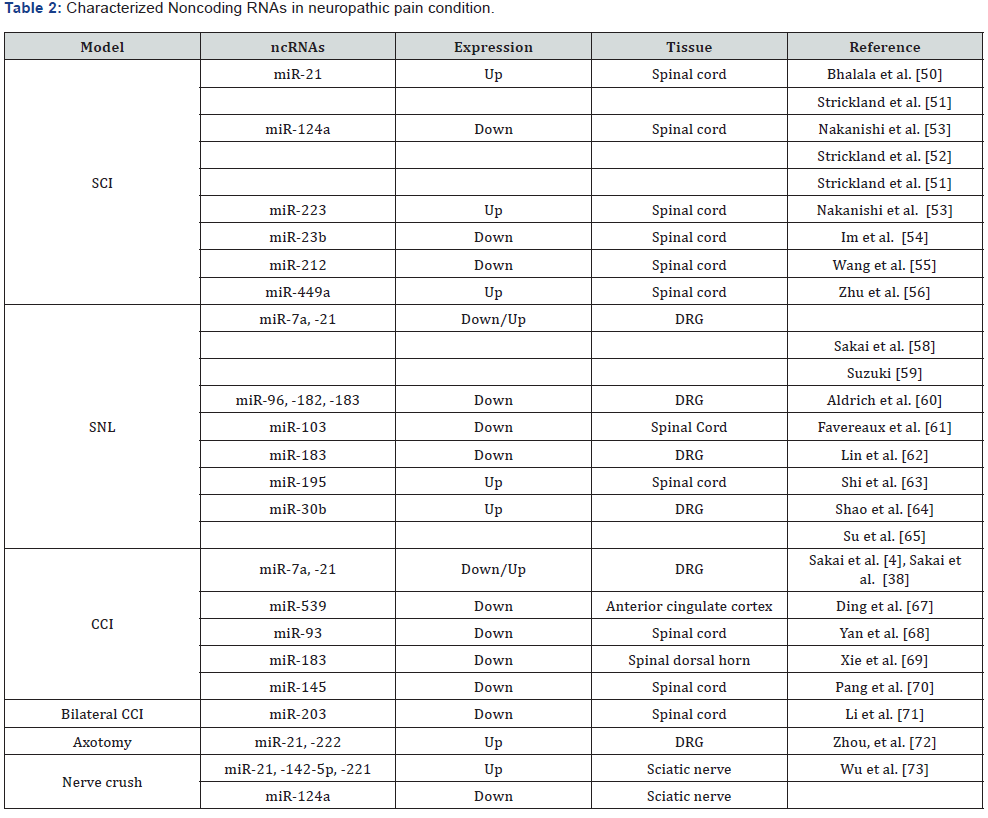
NcRNA (Noncoding RNA), SCI (Spinal cord injury), SNL (Spinal nerve ligation), CCI (Chronic constriction injury).
To start explaining the role of miRNA in neuropathic pain,
let`s first mention some of the main animal models that have
been development in this area. First the spinal cord injury (SCI)
was proposed by Allen AR in 1911 [47] then in [48], adapted the
Allen`s method by a briefly laminectomy performed at the T9–
10 thoracic vertebrae level to expose the spinal cord at T10 and
inducing the SCI by New York University Impactor device [49],
with this methods it has been possible to correlate the injury
of the spinal cord with the regulation and expression of miRs
like miR-21, miR-124a, miR-23b, miR-223, miR-449a and miR-
212 [50-56]. Spinal nerve ligation (SNL), the left L6 transverse
process is removed to expose the L4 and L5 spinal nerves then
the L5 spinal nerve is carefully isolated, tightly ligated with 3-0
silk thread, and transected just distal to the ligature [57], with
this method it`s has been studied the miRs miR-7a, -21, -96,
-182, -183, -103, -195 [58-63] and more recently with miARN-
30b [64,65]. The chronic constriction injury (CCI) model was
proposed by Bennett and Xie in 1988 [66], in this model the right
sciatic nerve is tied loosely with four ligatures by chromic cat gut
4-0, the lastly works with this method revealed the expression
of miR-7a, -21, -539, -93, -183, -145 and -203 [58,59,67-71].
The axotomy model consist in a transection of the sciatic nerve
at a point approximately 1 cm distal to the exit point of spinal
nerve roots, after Axotomy the expression of miR-21 and miR-
222 increased in DRG [72]. Finally, the Nerve crush model is
achieved after expose sciatic nerve and crush in the mid-thigh
for 15sec with a fine hemostat, in the day 4 and 7 post injury
the three most highly up regulated miRNAs was miR-21, miR-
142-5p, and miR-221 [73]. Now we`ll mention some of the most
representative and lastly found miR`s involved in neuropathic
pain development.
The miR-21 is expressed in all the neuropathic pain models,
[54] demonstrated that miR-21 transcripts are physiologically
regulated by peripheral nerve injury. Their role appeared to
be enhance neurite outgrowth from DRG neurons by targeting the Sprouty2 protein (SPRY2) 3′ UTR region in rats after
axotomy. More recently, [69] studied the role of miR-183 in
the development of neuropathic pain using the CCI model
they revealed that miR-183 can suppress AMPA receptors by
inhibiting the mammalian target of rapamycin (mTOR)/ vascular
endothelial growth factor (VEGF) pathway, which alleviates the
mechanical hypersensitivity associated with inflammation and
neuropathy [74]. Shao et al. [64] evidenced that one of the major
targets in neuropathic pain, the voltage-gated sodium channel
Nav1.7 are directly target by miR-30b. The expression of Nav
1.7 increases in nociceptive neurons during the development of
inflammatory hyperalgesia, while the knockdown or ablation of
Nav1.7 expression relieves inflammatory pain and hyperalgesia
[75]. Finally, [70] study suggested that miR-145 serves an
important role in the development of neuropathic pain through
regulating RREB1 expression and the PI3K/AKT signaling
pathway which serves an important role in vascular endothelial
growth factor (VEGF)-induced hyperalgesia [76].
Future Approaches and Conclusion
The studies reviewed in this article may us consider the
microRNA`s as potential targets and biomarkers for prediction
and treatment of several pain conditions. Because of their
role as master switches in regulation and signaling pathways
through modifications in nociceptive receptors, ion channels,
pro-inflammatory molecules, emotional and cognitional
behaviors associated with pain, the triggered enthusiasm for
miRNAs as promising therapeutic targets is still active. However,
challenges with respect to the use of miRNA-based therapeutics
in humans remain to be further explored [77]. When we can
fully understand the role of miRNAs in pain mechanisms, it will
be possible to maximize miRNAs potency while minimizing off
target toxicity and immunogenicity to provide great benefit for
clinical diagnostic and therapeutic applications.
Acknowledgement
Dr. Carlos H. Laino is greatly acknowledged for his help in the
critical review of this work.
For
more Open access journals Publishers Please visit our website: Juniper publishers




Comments
Post a Comment