MazEF homologous Modules System and A Post-segregational killing Mechanism (Bacteriostatic & Bactericidal Mechanism) of Novel Compound Isolated from Spondias monbinon Escherichia Coli and Bacillus Subtilis- Juniper Publishers
Juniper Publishers- Journal of Cell Science
Abstract
The basic objective of this research work is to evaluate the mechanism of action of compound Epigallocatechin, Epicatechin and Stigmasterol Phytosterol (Synergy), Aspidofractinine-3-methanol) and Terephthalic acid, dodecyl 2-ethylhexyl ester with Selected clinical isolates by using molecular biomarker MazEF9 TA system. Toxin-Antitoxin (TA) systems are highly conserved in members of the Gram positive +ve and Gram negative –ve bacteria which has been proposed to play an important role in physiology and virulence. Clinical microorganisms were cultured and Sub-culturing in Department of Microbiology and Centre for Biocomputing and Drug Development (CBDD), Adekunle Ajasin University, Akungba Akoko, Ondo-state, Nigeria. A 12 hours old culture of each microorganism was re-suspended in plant extract at 1000 μg mL in a total volume of 500μl for 0, 15, 30, 45, 60, and 180 minutes.
Keywords: Post segregational killing mechanism epigallocatechin; Epicatechin; Stigmasterol phytosterol; Mazef TA System; Bacteriostatic
Go to
Introduction
Spondiasmombin L. (Anacardiacaea) also known as hog plum is a plant that grows in almost every part of the world. It is fruit feriousdecidous plant of about 20m high that grows in the rain forest and the coastal area of Africa. It is known locally as “iyeye” by the Yoruba people of Nigeria Ripped fruits are eaten out hand by the old and young and processed into ice-cream, cool beverages, wine, jam and other preservatives. Spondiasmombin alsofound application in folk medicine. Infusion of its leaves has been used for a long time, without any report of collateral effect due to its anti- vitrotic activity against the herpes virus [1]. A tea of the flowers and the leaves is taken to relieve stomachache biliousness, urethritis, cystitis and eye and throat inflammation. Herbalist in South Western Nigeria use the plant in the treatment of typhoid, tuberculosis, diabetics, nervous disorders and psychiatric disorders. Offiah, Anyanwu [2] have reported the abortifacient activity of the aqueous leaf extract of Spondiamombin. In addition, the anthelmintic, molluscicidal, anxiolytic, anti-bacteria, antiviral effect of the plant have been previously described. Idu et al. [3] reported the inhibitory activity of Spondiamombin against Cycasrevoluta induced carcinogenesis. The discovery of toxin-antitoxin gene pairs (also called homologous modules) on extra-chromosomal elements of Escherichia coli, Bacillus subtilis and particularly the discovery of homologous modules on the bacterial chromosome, suggest that a potential for programmed cell death may be inherent in bacterial cultures which is under the scope of this research work, this was reported on the E. coli / Bacillus subtilis mazEF system, a regulatable addiction module located on the bacterial chromosome. MazF is a stable toxin and a labile antitoxin. it show that cell death mediated by the E. coli /Bacillus subtilis mazEF module can be triggered by several medicinal plant extract and antibiotics like Rifampicin, chloramphenicol, and spectinomycinetc, that are general inhibitors of transcription and translation. These medicinal plant extract and antibiotics alike inhibit the continuous expression of the labile antitoxin MazE, and as a result, the stable toxin MazF causes bacteria cell death. The results of this research have implications for the possible mode(s) of action of this group of medicinal plant and antibiotics on selected clinical isolates. The mode of action of the medicinal plants may be either bacteriostatic or bacteriocide [4] but the scope of this research work is based on bacteriostatic mechanism of action of isolated novel compound on selected clinical organisms (E. coli and B. subtilis).
In Escherichia coli/Bacillus subtilis cultures, programmed cell death is mediated through a unique genetic system. This system, called “homologous module,” consists of a pair of genes that specify for two components: a stable toxin and an unstable antitoxin which prevents the lethal action of the toxin. Until recently, such genetic systems for bacterial programmed cell death have been found mainly in E. coli and Bacillus subtilis on low-copy-number plasmids, where they are responsible for what is called the postsegregational killing mechanism. When bacteria lose the plasmid( s) (or other extra-chromosomal elements), the cured cells are selectively killed because the unstable antitoxin is degraded faster than is the more stable toxin [5]. Thus, the cells are “addicted homologous” to the short-lived product, since its de novo synthesis is essential for cell survival [6].Therefore, these homologous modules have been implicated as having a role in maintaining stability in the host of the extrachromosomal elements on which they are bounded, Jensen, Gerdes [7], once this stability is destroyed, this will inevitably leads to the death of the bacterial cell. Pairs of genes homologous to some of these extra-chromosomal addiction modules have been found on the E. coli and Bacillus subtilis chromosome Aizenman et al. [8], Gerdes et al. [9]. The mazEF module consists of two adjacent genes, mazE and mazF, located in the rel operon downstream from the relA gene Metzger et al. [10].
In the study, mazEF was found to have the properties required for an addiction module:
(i) MazF is toxic and MazE is antitoxic
(ii) MazF is long lived, while MazE is a labile protein degraded in vivo by the ATP-dependent ClpPA serine protease
(iii) MazE and MazF interact
(iv) MazE and MazF are coexpressed
Both MazF and MazE are expressed thereby leading overexpressed killing effects of bacteria cell, Moreover, the mazEF system has a unique property: its expression is inhibited by guanosine 3′,5′-bispyrophosphate (ppGpp), which is synthesized under conditions of extreme amino acid starvation by the RelA protein [11] thereby leading to the death of the bacteria cell. Based on these properties of mazEF and on the requirement for the continuous expression of MazE to program cell death, members of our group offered a model for programmed cell death under conditions of nutrient starvation [8]. In this study, MazF (toxic) is used to stimulate the programme cell death of the clinical isolates and evaluates the efficacy of novel compound isolated from Spondiasmombn plant on the selected clinical organisms.
Go to
Materials and Method
Microorganism for the research work
Clinical microorganisms were used for this research work, which comprised Escherichia coli ATCC 25922and Bacillus subtilis.
Sources of microorganisms
All typed strains used for this research work were purchased from the University of Pennsylvania, School of Medicine, Philadelphia, United States of America (USA), in an America Type Culture Collection (ATCC) , and the other locally isolated bacteria and fungi were clinical organisms collected from Central Medical Laboratory( CML), Obafemi Awolowo University Teaching, Hospital (OAUTH), Ile Ife, Osun State, and the Institute of Advance Medical Research and Training (IMRAT),University College Hospital, Ibadan, Oyo State Nigeria.
Authentication of test microorganisms
The identity of the test organisms was confirmed using Biomerieux, France, API 20E Kits for bacteria as specified by the manufacturer’s instruction. Analytical Profile Index [12].
Isolation of RNA.
A 12 hours old culture of each microorganism was re-suspended in plant extract at 1000μg mL in a total volume of 500μl for 0, 15, 30, 45, 60, and 180 minutes. The cells were pelleted by centrifugation at 5000g for 5 minutes. The pellets were rinsed twice in phosphate buffer saline (PBS). Then 1/10 volume of 95% ethanol plus 5% saturated phenol were added to the pellets to stabilise cellular RNA. The cells were then re-harvested by centrifugation (8200g, 4°C and 2 minutes). The supernatant was aspirated and pellets re-suspended in 800 μl of lysis buffer (10mMTris, adjusted to pH 8.0 with HCl, 1mM EDTA) and 8.3 U/ml Ready-LyseTM Lysozyme Solution. After the pellets were re-suspended, 80μl of a 10% SDS solution was added, mixed and incubated for 2 minutes at 64°C. Then 88μl of 1 M NaOAc (pH 5.2) was mixed with the lysate followed by an equal volume of water and saturated phenol was added. This was incubated at 64°C for 6 minutes while inverting the tubes every 40 seconds. The aqueous phase was separated following centrifugation at 21,000g for 10 minutes at 4 °C. The RNA was precipitated from the aqueous layer using 1/10 volume of 3M NaOAc (pH 5.2), 1mM EDTA and 2 volumes cold EtOH and centrifugation at 21,000g for 25 minutes at 4°C. Pellets were washed with ice cold 80% EtOH and centrifuged at 21,000g for 5 minutes at 4°C. The ethanol was carefully removed and the pellets were air dried for 20 minutes. The pellets from each split
Solation of RNA from bacterial cell
Figure 1.
PCR protocol
Reverse Transcription–PCR reaction was performed in a 15.0μl final volume. Briefly, 1μl template cDNA (~40 ng) was combined with 1.0μl of forward primer (5nM), 1.0μl of reverse primer(5nM), 4.5 ml nuclease-free water and 7.5 μl of Taq 2X Master Mix. Thermo cycling was performed by 40 cycles at 95°C for 15 seconds, 60 °C for 15 seconds and 72°C for 15 seconds. Analysis of the PCR products was performed using 1.5% agarose gel solution in TBE buffer and visualisation was enabled by soaking gel in ethidium bromide solution for 10 minutes and UV-transilluminator. The data obtained were analyed using Graph pad prism version 6.01 description and frequency. statistic was generated to describe the diameter of inhibition. quantitative phytochemical constituent and toxicological prameter to test for the level of significance [1].
Gel electrophoresis
Assessment of Polymerase Chain Reaction products (amplicons) were electrophoresed in 0.5% of agarose gel using 0.5×TBE buffer (2.6g of Tris base, 5g of Tris boric acid and 2 ml of 0.5M EDTA and adjusted to pH 8.3 with the sodium hydroxide pellet) with 0.5μ lethidum bromide. The expression product was visualized as bands by UV-transilluminator [1] Table 1.
Go to
Results
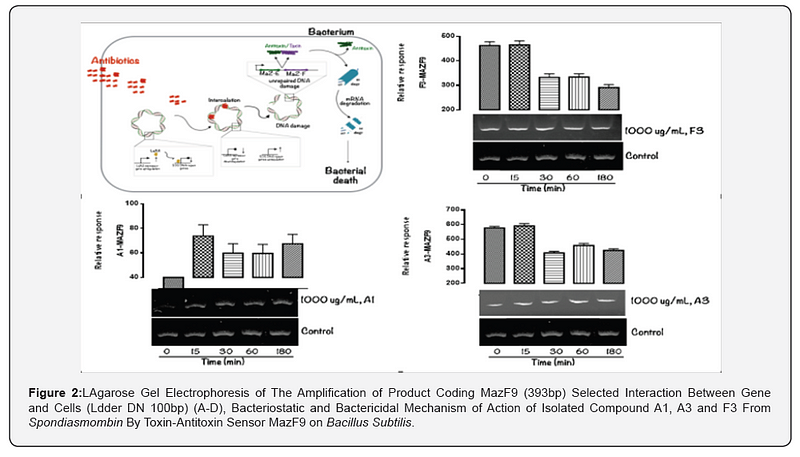
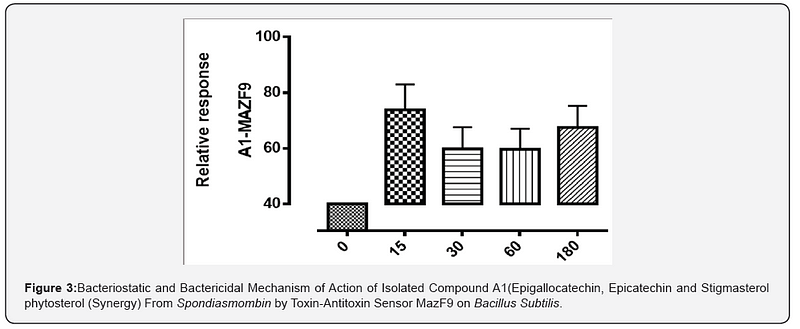
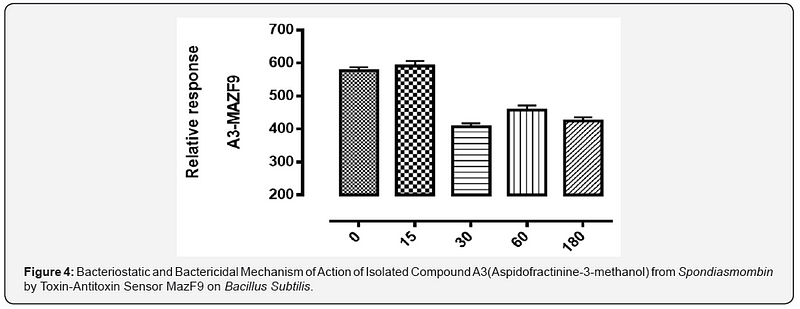
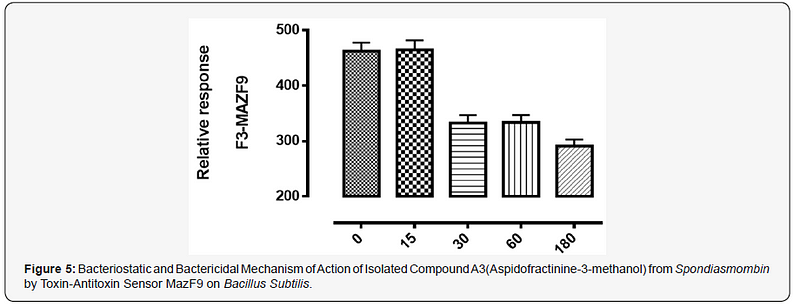
The MazEFhomologous modules system and a Post-segregational killing mechanism (Bacteriostatic & Bactericidal mechanism) of novel compound isolated from Spondiasmonbinon Escherichia coli and Bacillus subtilis were demonstrated in Figure 1–6. The novel compound isolate from Spondiasmombin include the following, (A3-Epigallocatechin, Epicatechin and Stigmasterol phytosterol (synergy), A3-Aspidofractinine-3-methanol) and F3(Terephthalic acid, dodecyl 2-ethylhexyl ester) and the gene expression MazF9was observed in Escherichia coli and bacillus subtilis. Figure 1 shows Bacillus subtilis, mazEF homologous modules system and Post-segregational killing mechanism (Bacteriostatic & Bactericidal mechanism) of compounds A1(Epigallocatechin, Epicatechin and Stigmasterol Phytosterol (Synergy), A3(Aspidofractinine- 3-methanol) and F3 (Terephthalic dodecyl 2-ethylhexyl ester) by MazEF9 (Toxin/ antitoxin sensor. In Figure 2,3 and 4. It was observed that compounds A1, A3 and F3 have a deleterious effect on the cell (via DNA) of the test organism. At 180 minutes, the death phase was ascertained. This is graphically demonstrated in the figure1. Both MazE (toxin) and MazF (antitoxin) were produced, which both were used to activate the death phase of the cell through Post-segregational killing mechanism (Bacteriostatic & Bactericidal mechanism) of MazEFhomologous modules system on E. coli and B. Subtilis. The compounds have bacteriostatic and bactericidal effect on the test organisms (E. coli and B. subtilis). Figure 5 shows E.coli, mazEF homologous modules system and Post-segregational killing mechanism (Bacteriostatic & Bactericidal mechanism)of A1(Epigallocatechin, Epicatechin and Stigmasterol Phytosterol (Synergy),A3 (Aspidofractinine-3-methanol) and F3 (Terephthalic dodecyl 2-ethylhexyl ester)by gene expression MazEF9 (Toxin/ antitoxin sensor ) bacteriostatic and bactericidal effect as the mechanism of action compounds of A1, A3 and F3 as observed in (Figure 6–8) respectively.
Go to
Discussion
The purpose of the research work is to determine the Post-segregational killing mechanism (Bacteriostatic & Bactericidal mechanism) of novel compound isolated from Spondiasmonbinon Escherichia coli and Bacillus subtilis by MazEFhomologous modules system. action of compound A1 (Epigallocatechin, Epicatechin and Stigmasterol Phytosterol (Synergy), A3(Aspidofractinine-3-methanol) and F3(Terephthalic acid, dodecyl 2-ethylhexyl ester) with two Selected microorganism (Gram positive +ve), (Gram negative –ve) were demonstrated by toxin-antitoxin gene pairs (also called addiction modules) on extra-chromosomal elements of Escherichia coli, Bacillus subtilis All figs describe the mechanism of action of the isolated compound of ethyl acetate stem bark extract of Spondiasmombin extract. Nine of these TA systems belong to the mazEF family, encoding the intracellular MazF toxin and its antitoxin. Another mechanism of action of compound A1, A3 and F3 on the selected microbe E. coli and Bacillus subtilis is by probing for the toxin-antitoxin Maz F9 bio sensor. It was observed that in Figure 1–3. A1 MazF9, A3Maz F9 and F3 MazF9 stimulate the production of toxin/antitoxin system in the cell of the microorganism, used for this study and a greater effect were found between 0 to 15 minutes and decrease in effect were found at 30 to 180 minutes, this effect of toxin-antitoxin leads to the death of the organism between 0 to 180 minutes, the death ratio is measured and cells were destroyed or damaged by stimulation of toxin-antitoxin system. Toxin-antitoxin system are induced to measure the death phase at 0–180 minutes and this study was conducted to investigate the role of MazF9 to isolated compound. It was also observed that over expression of MazF9 induced a state of irreversible bacteriostasis consistent with the results of the previous study [13- 15]. The main finding of this work is that E. coli mazEF-mediated cell death can be triggered by medicinal plant that are general inhibitors of transcription and/or translation. it was shown that these Spondiasmombin reduce the cellular level of the antitoxic labile protein MazE and seem thereby to permit the lethal action of the toxic protein MazF. The effect of the Spondiasmombin both on cell death and on the reduction in the cellular level of MazE [16].
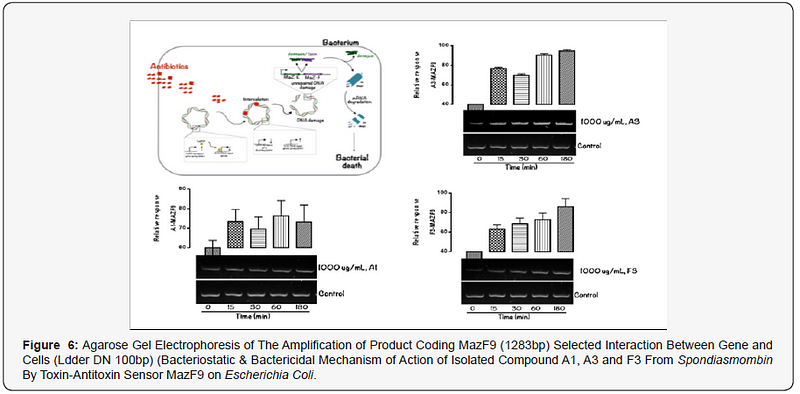
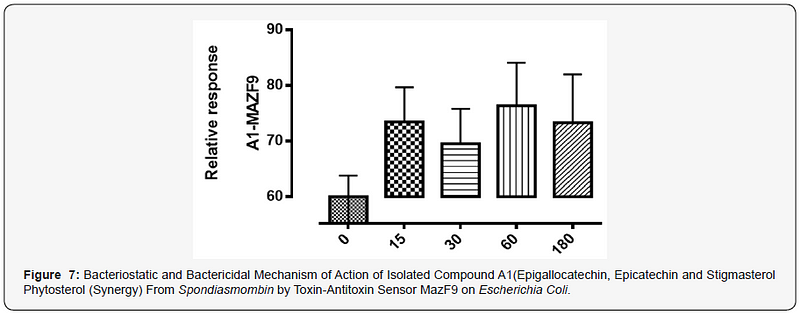
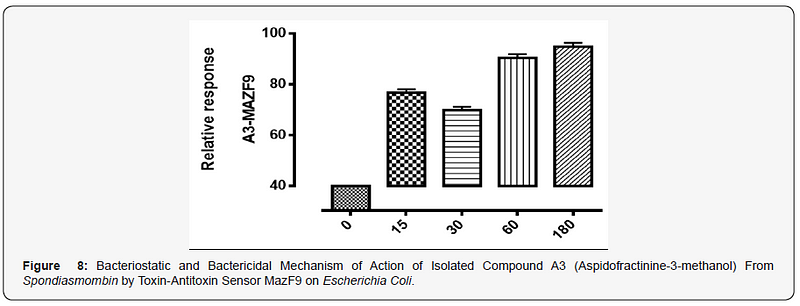
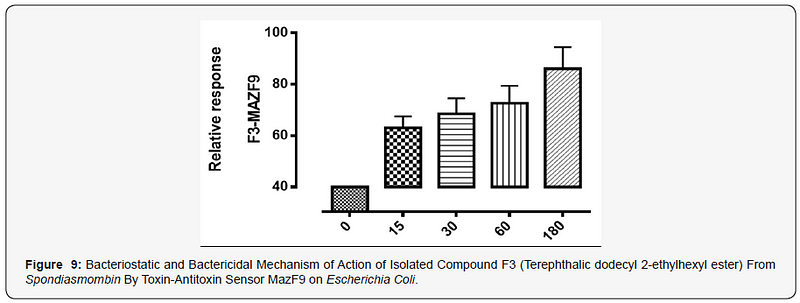
The isolated compound has a great effect on selected organism. It was observed that the effect of gene MazF9 has more lethal effects in E. coli, taking a cognizant from figure 5–8 represented by A1MazF9, A3MazF9 and F3MazF9. It was observed that the compound complete inhibits the E. coli at 180 minutes i.e. the compound has a better activity on E. coli than B. subtilis. The reasons for this activity must be as a result of their cell wall, the Gram negative (E. coli) has a greater activity because of a thin composition of the peptidoglycan cell wall compares to the gram positive (B. subtilis) which has a large composition of peptidoglycan in the cell wall. The gram negative is easily affected by antibiotic compound and compares the gram positive with very thick cell wall, this account for the activity of MazF9 more on E. coli than B. subtilis but their death rate was constantly measured, and toxin-antitoxin activity were clearly depleted [17].
In analogy to the programmed cell death apparatus in eukaryotic cells [18] reported that the suicide machinery in bacterial cells is always present, it is only requires a trigger to activate it. Moreover, at least for E. coli, the straightforward choice is death caused by a stable intracellular toxin (in this case MazF). The choice of life over the “default” death requires a dynamic antagonistic process manifested either by the continued production of the unstable antitoxin (in this case MazE) or by a process that would prevent the degradation of the unstable antitoxin. Thus, cell death could be caused by anything that would prevent the continuous expression of the antitoxic protein MazE [4]. It should be noted that toxin/ antitoxin system is widespread in bacterial genomes and contribute to prokaryotic stress adaptation and the formation of persister cells biofilms [19]. The mechanism of toxin components is to exert their effects in different ways by targeting essential cellular functions such as DNA replication, protein synthesis, cell division, peptidoglycan biosynthesis (see above) and ribosome assembly.
However, RNA cleavage is the most prevalent mode of action in the pathway [20]. Vasperet al., [21]reported that in E.coli (see above), one of the most well characterized toxin-antoxin gene is MazF9, an operan that encodes that intracellular MazF and its cognate antitoxin MazF9 toxin cleaves MRNA and DNA at 3,5 or 7 base recognition sequences in different bacteria and have been implicated in controlling cellular responses to various adverse conditions encountered by the bacteria in the host. Other mechanism of MazF9 in E. coli is MazF9 protein cleaves both MRNA and 16s ribosomal RNA and proposed to generate a subpopulation of stress ribosomes, thereby enabling the translation of leaderless transcript [22]. MazF9 system in E. coli are activated on exposure to numerous stress condition in an extracellular death factor-dependent manner. Another mechanism of action of compound A1, A3 and F3 on the selected microbes E. coli and Bacillus subtilis was demonstrated by probing for the toxin-antitoxin MazF9 gene. This effect of toxin-antitoxin MazF9 biosensor leads to the death of the organisms between 0 to 180 minutes. The compounds have more lethal effects as they completely inhibited the E. coli at 180 minutes. In addition, the compounds have a better activity on E. coli than B. subtilis. The reasons for this activity must be as a result of differences in their cell wall compositions.
This results in Figure 1–8, showing that the mazEF system is responsible for approximately 90% killing by Spondiasmombin and other medicinal plant and antibiotics alike may illuminate, the until now elusive cause of E. coli killing by medicinal plant. Furthermore, medicinal plant like Spondiasmombin are considered to be bacteriostatic and bactericidal in action [23]. It should be noted that toxin system is widespread in bacterial genomes and contribute to prokaryotic stress adaptation and the formation of persister cells biofilms [19]. The mechanism of toxin components is to exert their effects in different ways by targeting essential cellular functions such as DNA replication, protein synthesis, cell division, peptidoglycan biosynthesis (see above) and ribosome assembly. The mechanism of action of toxin-antitoxin gene MazF9 include RNA cleavage [20]., MRNA and DNA cleavage and protein cleavage of both MRNA and 16s ribosomal RNA [22].
Go to
Conclusion
This is better method of measuring the mechanism of action of novel compound by triggering the toxin/antitoxin cell death of inherent nature of bacterial cell.
For more Open access journals
please visit our site: Juniper Publishers
For more articles please click
on Journal of Cell Science & Molecular
Biology




Comments
Post a Comment